Using Python on M2
In another post, I mentioned separating my personal computer environment from the company's one when I bought Miorine (my M2 Max MBP). I also set up my private development environment with Neovim because I thought using my company's JetBrains licenses on my personal laptop was inappropriate. It was interesting because it was my first time (after 30 years since I learned vi in college) readily accepting vi/vim for my main flow. And there was a small challenge yesterday when I decided to set up a Python environment.
I haven't used Python even in my work because 1) the primary language for most of my tasks is Go, and (moreover) 2) a psychological barrier. I should have used Python 3.7 for work as there are some dependencies (as usual), but the problem was that the Apple Silicon environment didn't support Python 3.7; that means I had to use Rosetta (and Rosetta iTerm terminal), which I wasn't up to. So, I had put some distance to Python at work - anyhow, I haven't had any tasks that required Python. However, at this right moment, when everything was getting back to normal, I got my first COVID and was isolated in my bedroom to protect other family members. I needed something to coax myself and started watching ML and data processing (e.g., Pandas, Numpy, TensorFlow, etc.) lectures on YouTube. I wanted to try some sample code, and the fact that I didn't need to be bothered by a specific Python version and Rosetta at home invigorated me. So I decided to set up a genuine (Apple Silicon native, and based on Neovim) Python development environment on my M2 Max MBP.
Installing Python
Let's get it straight; I don't like Python. There would be many reasons, but the most significant one is there are too many ways to do something. Just installing M1/M2 native Python could be done in many ways. Apple has its own guide, but I found that it produced some warnings that made me a little reluctant to use it. After some investigations, my choice was to use miniforge to install Conda and manage the Python version and environment: creating a Conda environment and using one Python version for it. Miniforge? Conda? Yes, it's confusing; as I said, that's why I don't like the Python environment. We need to clarify some concepts.
- Conda is an open-source package and environment management system: a package management system such as pip and an environment management system like venv - simultaneously.
- Anaconda is a set of hundreds of packages, including Conda, Numpy, etc. The catchphrase of Anaconda is "Where packages, notebooks, projects, and environments are shared." It is also worth to note Anaconda is a private company.
- Miniconda is a small subset of Anaconda that only has Conda and its basic dependencies.
- Conda-forge is a GitHub community-driven organization containing repositories of Conda recipes. You can think of it as an alternative channel for package installations.
- Miniforge (finally!) is a GitHub repository and a community (Conda-forge) driven minimal Conda installer.
While Anaconda and Miniconda did not support the Apple Silicon environment (although now Miniconda supports arm64), Miniforge provided an installer for arm64 (Apple Silicon) architecture; that's why I was going to use Miniforge.
Download the Miniforge3 arm64 installer script from the miniforge git repository.
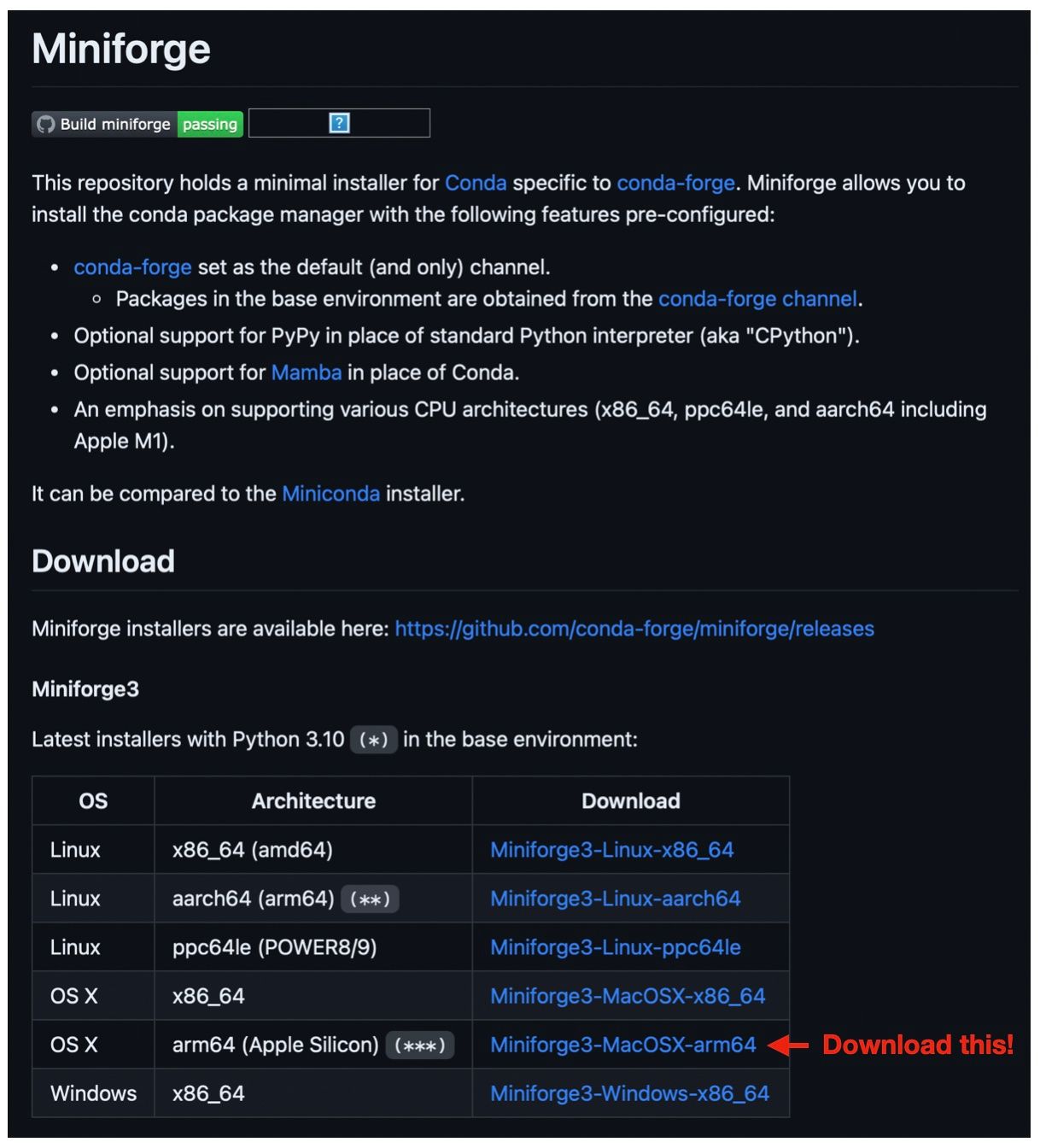
Run the script. It will install arm64 Conda and Python 3.10.10 (the latest one).
sh ~/Downloads/Miniforge3-MacOSX-arm64.sh
Once it is installed and we rerun the terminal, it will automatically activate the Conda base environment. You can install any additional packages (e.g., pandas) either using pip install or conda install.
Some guides had used Pyenv for multiple Python versions, but I thought it was unnecessary (at least for me).
Additionally, you can install TensorFlow for macOS (using Apple Silicon) and the tensorflor-metal plug-in.
pip install tensorflow-macos
pip install tensorflow-metalLastly, I verified installations with the following script as noted in Apple's guide.
import tensorflow as tf
cifar = tf.keras.datasets.cifar100
(x_train, y_train), (x_test, y_test) = cifar.load_data()
model = tf.keras.applications.ResNet50(
include_top=True,
weights=None,
input_shape=(32, 32, 3),
classes=100,)
loss_fn = tf.keras.losses.SparseCategoricalCrossentropy(from_logits=True)
model.compile(optimizer="adam", loss=loss_fn, metrics=["accuracy"])
model.fit(x_train, y_train, epochs=5, batch_size=64)
Everything seemed to be perfect. However, the trouble came with the Neovim.
Neovim for Python
I had set up pyright for the Python LSP server. So, I expected it would work seamlessly. What I don't like about the modern GUI IDE is that it usually has its own execution environment. Therefore, we have to set up a new (different from the existing terminal) environment for the IDE; if I cannot build the source code while I can build it in the terminal, that's the typical implication of these separate environments. As Neovim (vim) runs in the terminal environment that it is launched from, they share the environment; we don't need to set up another environment for it.
It should have been... So, I was surprised when I typed the first line of a Pandas sample code.
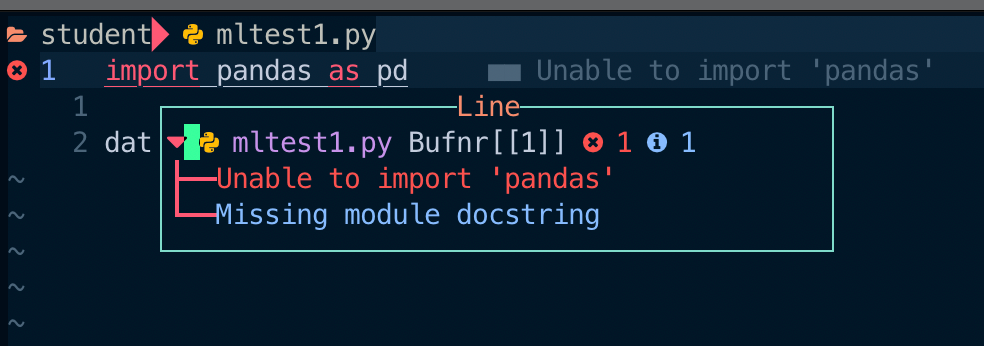
What?... Neovim (precisely, pyright) couldn't resolve the pandas package? I checked the installed packages with the conda list command, and it (pandas) was definitely there.

At first, I guessed that pyright couldn't resolve the current (base) Conda environment. I created a pyrightconfig.json file and tried to specify stubPath, venvPath, and so on; nothing worked. I checked several Reddit and StackOverflow articles. I've come across a few similar cases, but unfortunately, none of them were of much help. It's frustrating to see that something should work smoothly, yet it doesn't. This is a real source of agony for developers. I tried several other Python LSP servers, such as the jedi-language-server and the anakin-language-server (what???). However, it didn't make any difference. Wait? no differences at all?
I tried pyright via the command line. It simply showed clean results; pyright had worked.

I checked the current file's LSP status with the LspInfo command.
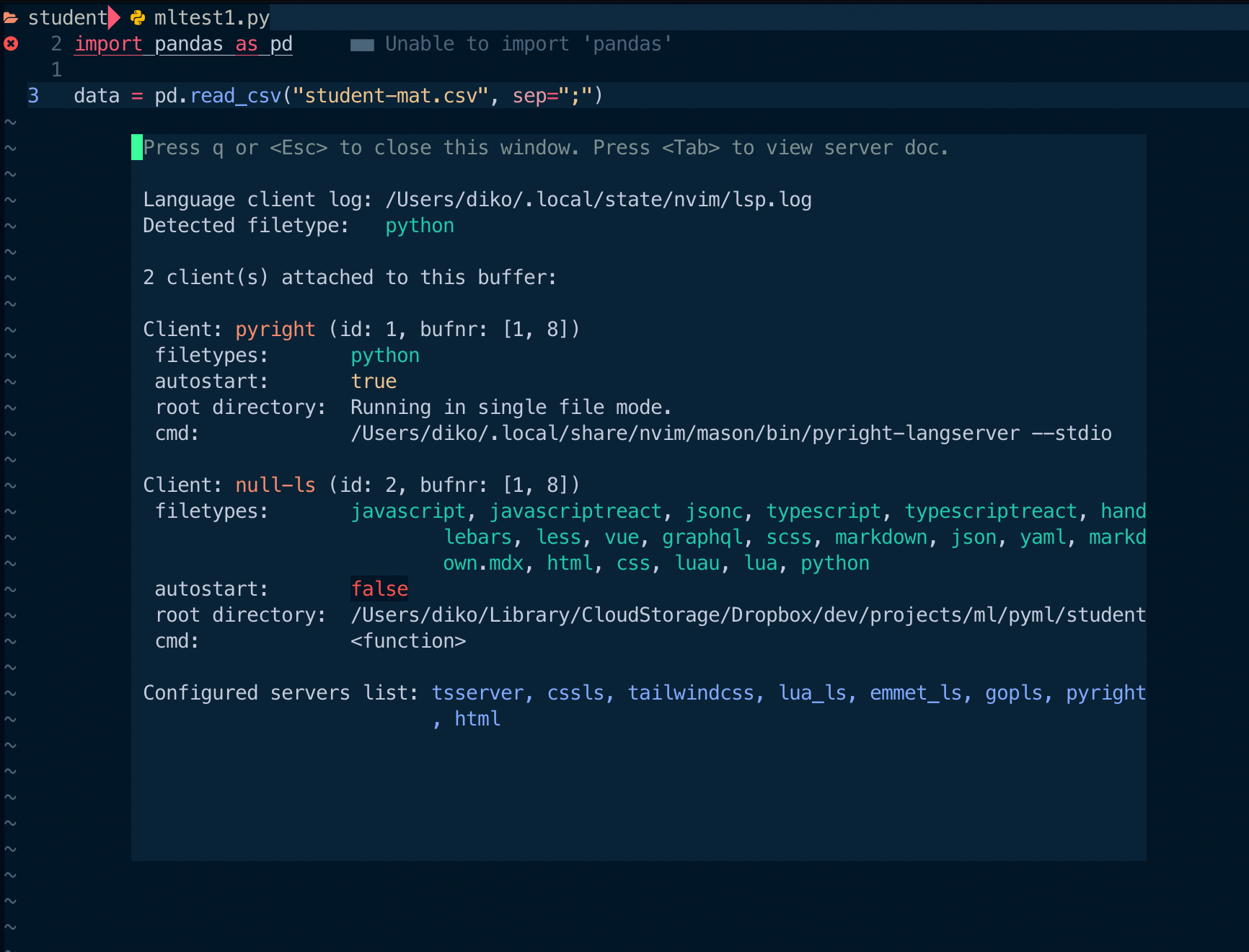
Ah-ha. pyright wasn't the only one involved in the diagnostics. Null-ls was another player behind the scene. It should have come along with a soullessly copied config file. I disabled Python diagnostics from null-ls.
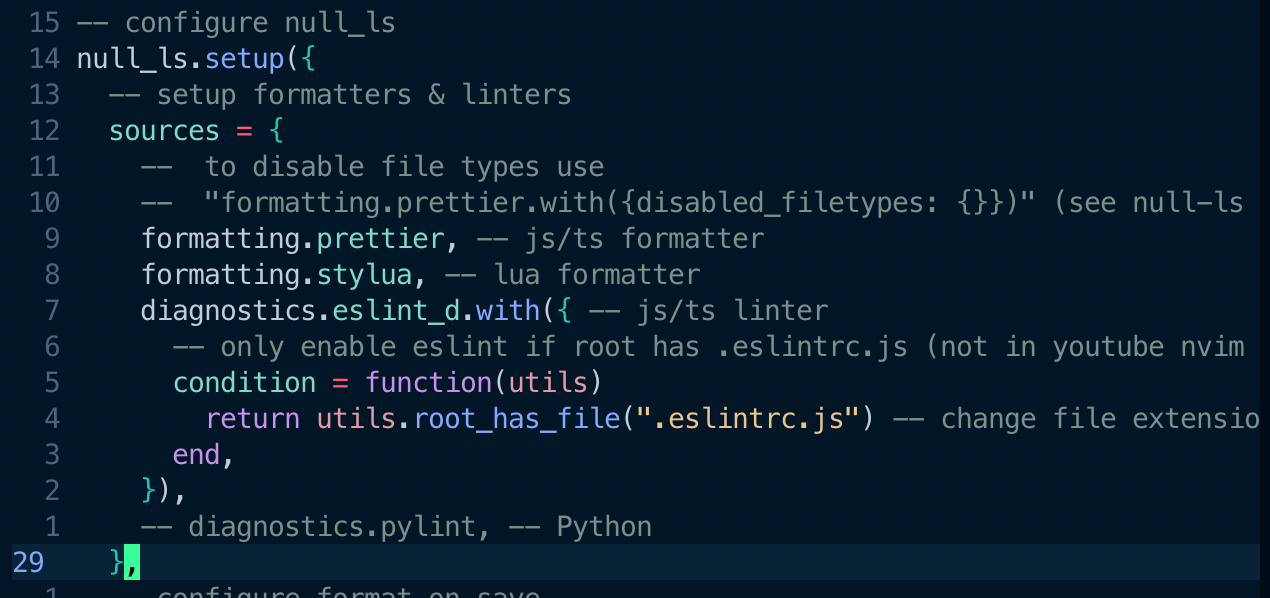
Bingo. The error was gone — a piece of mind.
If I had only merely copied configs and used features without understanding the internals, I couldn't resolve it and should get back to PyCharm. As an engineer, finding solutions by thoroughly comprehending the underlying systems should be a source of immense enjoyment. Truly it is.



